#MeetTheScientist
Esther Dorcely

My name is Esther Dorcely, and I studied Biomedical Sciences and Enterprise at Georgia State University. I discovered this study through my deep interest in science and medicine, an interest that’s been a part of me for as long as I can remember. My mother has been a nurse for over 30 years, and many of my close relatives have also pursued careers in healthcare. From a young age, they encouraged me to become a doctor or nurse. While both paths appealed to me, I found myself particularly drawn to becoming a Pharmacist. That passion began early in life. I used to approach pharmacists with questions about their work what they loved most about it and how they got started. Outside of those conversations, I was constantly reading books about epidemics and diseases and watching documentaries on medical research and innovation. All of that exposure helped shape my vision for the future. Seeing healthcare professionals regardless of their specialty dedicate themselves to helping others truly inspired me. Science and medicine have come a long way, improving and saving countless lives. My goal is not only to help others but to also advocate for their health and wellness. As a minority, I understand the importance of representation and want to ensure that underrepresented communities receive the care and support they deserve.
Bridging Biology and Innovation.
E.Coli Testing (Allora Test Kits)
E.Coli Fimbrae (Indigo Instruments)
Break down
Research

Research Topic
What is E.Coli?
Escherichia coli (E. coli) is a bacterial organism that naturally resides in the intestinal tracts of humans and animals. Contrary to popular belief, most strains of E. coli are harmless and play a critical role in physiological functions such as digestion and vitamin production (E. coli Infection). However, E. coli can become pathogenic when introduced into the body through contaminated food sources or contact with contaminated surfaces,especially in healthcare settings. Pathogenic strains of E. coli can cause severe infections. For example, if E. coli enters the urinary tract, it may result in urinary tract infections (UTIs) that could potentially lead to kidney damage or failure (Virulence factors and drug resistance in Escherichia coli ).
Overcoming Bacterial Defenses
In such cases, patients are typically treated with antibiotics to eliminate the infection. Recent research suggests that combination therapies, rather than monotherapies, are more effective at enhancing the immune system’s ability to combat bacterial infections. As a result, many healthcare professionals now incorporate adjuvant agents that enhance the activity of antibiotics into treatment regimens.These adjuvants may work by increasing antibiotic potency or by targeting bacterial mechanisms responsible for resistance.
Potent Resistance Mechanisms
-Efflux pumps, which expel antibiotic compounds from bacterial cells, lowering drug concentration and effectiveness
-Target protection, which blocks the antibiotic’s binding site, reducing efficacy
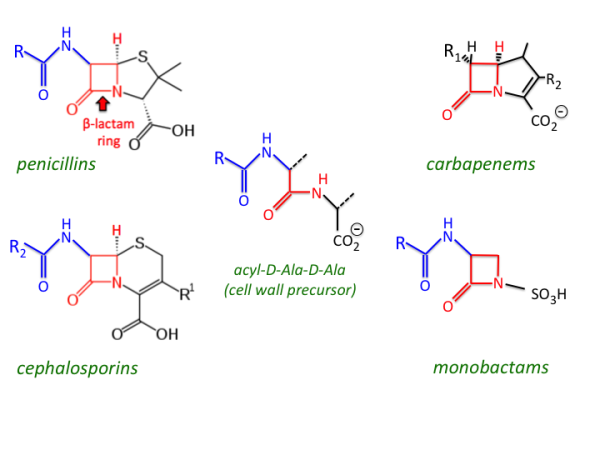
Enzymatic inactivation, where enzymes like beta-lactamases degrade antibiotic molecules, especially beta-lactam rings structures composed of three carbon atoms and one nitrogen atom that are critical to the antibiotic’s function
Beta-Lactamase-Driven Resistance in Bacterial Infections
Over time, antibiotic resistance has increasingly been observed in bacterial infections, including those caused by E. coli. Research has identified specific beta-lactamase enzymes in E. coli that contribute to this resistance. Two notable examples are blaTEM-1B and blaOXA-1, which encode TEM- and OXA-type beta-lactamases, respectively. These resistance genes are often located on plasmid small DNA molecules that facilitate the transfer and expression of antibiotic resistance traits between bacteria. TEM and OXA enzymes primarily function by hydrolyzing (breaking down) certain beta-lactam antibiotics, rendering them ineffective. The major challenge now lies in finding strategies to counteract the action of these enzymes. Emerging research highlights the potential of adjuvant antibiotics, which as previously mentioned can serve as powerful tools in overcoming beta-lactamase-mediated resistance and improving treatment outcomes.
What Can Be Done?
To address this challenge, scientists have developed effective strategies to control and, in some cases, halt the progression of antibiotic resistance. One such approach involves the combined use of beta-lactamase inhibitors and beta-lactam antimicrobials (Science Direct). This combination significantly reduces the potential for resistance to develop. The mechanism involves the inhibitors acting as substrates or reactants that bind to the beta-lactamase enzyme with high affinity. This binding disrupts the enzyme’s ability to function effectively, and in some cases, permanently inactivates it, greatly enhancing the efficacy of the antibiotic treatment.

Methodology
Comprehensive Antibiotic Resistance Database
My personal contribution involved the identification of beta-lactamase enzymes associated with the selected E. coli strain. To achieve this, I used the Comprehensive Antibiotic Resistance Database (CARD). CARD integrates the Antibiotic Resistance Ontology (ARO) with curated gene sequences and mutation data, enabling detailed analysis of antibiotic resistance within genomic and metagenomic datasets.
Within CARD, the Resistance Gene Identifier (RGI) tool was employed to predict antibiotic resistance genes based on genomic sequences. The RGI analysis produced multiple matches, including genes from resistance families such as OXA and TEM. The term “perfect hit” refers to a sequencing read that exactly matches a target sequence in the database or genome, indicating a high-confidence match. Of the fifteen perfect hits identified, three were beta-lactamase genes.
Mobile Element Finder
To gain a more comprehensive understanding of the genetic basis of these resistance mechanisms, it was necessary to determine which genes were responsible for encoding the identified enzymes. For this purpose, I used the Mobile Element Finder—a bioinformatics tool designed to detect segments of mobile DNA within the genome. These mobile elements play a key role in genetic adaptation, bacterial evolution, and the transmission of antibiotic resistance.
C.A.R.D. (ASM JOURNALS)